Overview
LINE (Large-scale Information Network Embedding) is a network embedding model that preserves the local or global network structures. LINE is able to scale to very large, arbitrary types of networks, it was originally proposed in 2015:
- J. Tang, M. Qu, M. Wang, M. Zhang, J. Yan, Q. Zhu, LINE: Large-scale Information Network Embedding (2015)
Concepts
First-order Proximity and Second-order Proximity
First-order proximity in a network shows the local proximity between two nodes, and it is contingent upon connectivity. A link between two nodes—or a stronger (positive) link weight—indicates higher first-order proximity. If no link exists between them, their first-order proximity is considered 0.
On the other hand, second-order proximity measures the similarity between the neighborhood structures of two nodes, based on shared neighbors. If two nodes have no common neighbors, their second-order proximity is 0.
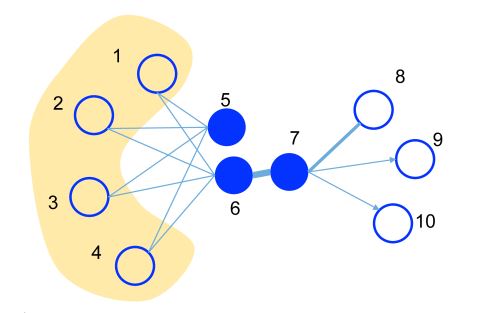
This is an illustrative example, where the edge thickness signifies its weight.
- A substantial weight on the edge between nodes 6 and 7 indicates a strong first-order proximity, meaning they should have similar representations in the embedding space.
- Although nodes 5 and 6 are not directly connected, their considerable common neighbors create a significant second-order proximity. As a result, they are also expected to be represented close to each other in the embedding space.
LINE Model
The LINE model is designed to embed nodes in graph G = (V,E) into low-dimensional vectors, while preserving either the first-order or second-order proximity between nodes.
LINE with First-order Proximity
To capture the first-order proximity, LINE defines the joint probability for each edge (i,j)∈E connecting nodes vi and vj as follows:
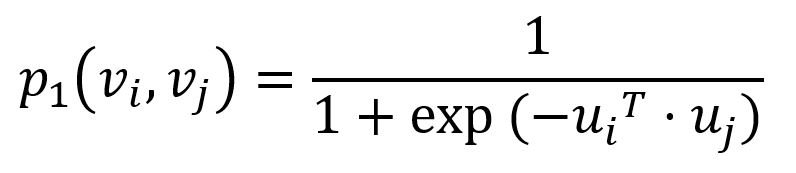
where ui is the low-dimensional vector representation of node vi. The joint probability p1 ranges from 0 to 1: the closer the vectors (i.e., the higher their dot product), the higher the joint probability.
Empirically, the joint probability between node vi and vj can be defined as
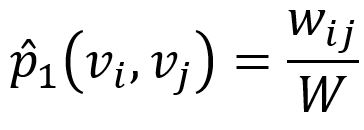
where wij denotes the edge weight between nodes vi and vj, W is the sum of all edge weights in the graph.
The KL-divergence is adopted to measure the difference between two distributions:
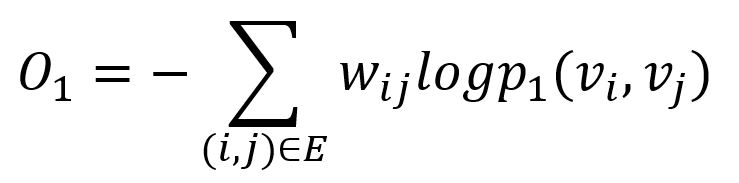
This serves as the objective function that needs to be minimized during training when preserving the first-order proximity.
LINE with Second-order Proximity
To model the second-order proximity, LINE defines two roles for each node - one as the node itself, another as "context" for other nodes (this concept originates from the Skip-gram model). Accordingly, two vector representations are introduced for each node.
For each edge (i,j)∈E, LINE defines the probability of "context" vj be observed by node vi as
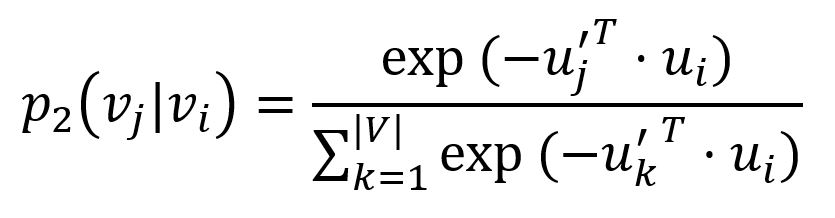
where u'j is the representation of node vj when it is regarded as the "context". Importantly, the denominator involves the whole "context" in the graph.
The corresponding empirical probability can be defined as
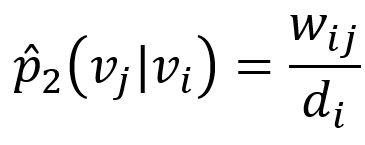
where wij is weight of edge (i,j), di is the weighted degree of node vi.
Similarly, the KL-divergence is adopted to measure the difference between two distributions:
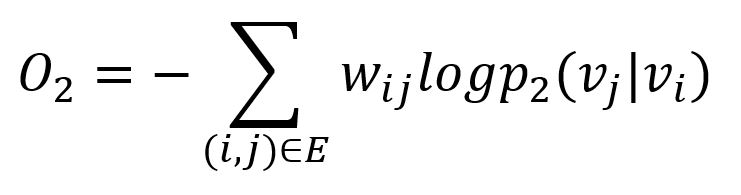
This serves as the objective function that needs to be minimized during training when preserving the second-order proximity.
Model Optimization
Negative Sampling
To improve computation efficiency, LINE uses negative sampling, drawing multiple negative edges from a noise distribution for each edge (i,j). Specifically, the two objective functions are adjusted as:
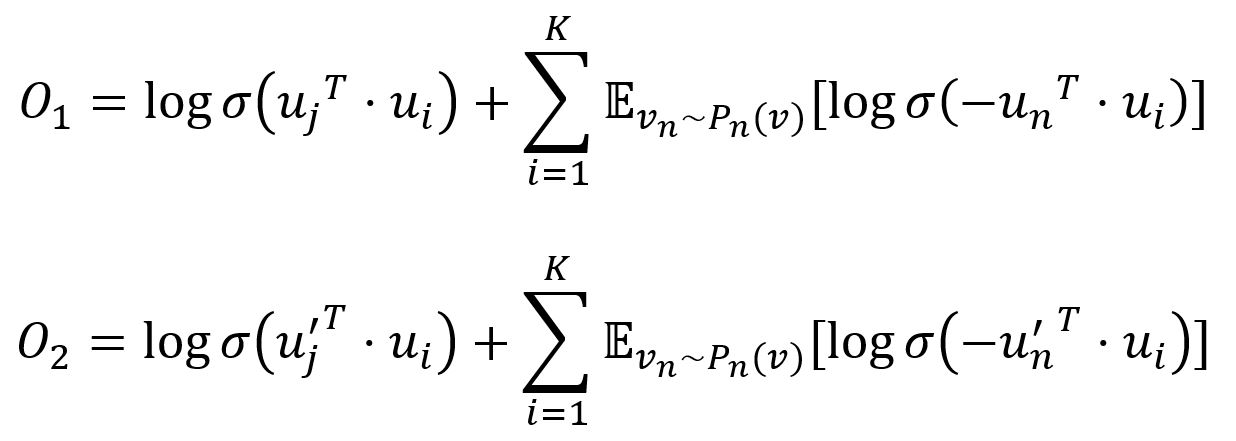
where σ is the sigmoid function, K is the number of negative edges drawn from the noise distribution Pn(v) ∝ dv3/4, dv is the weighted degree of node v.
Edge-Sampling
Since edge weights are included in both objective functions, they are also applied to the gradients during optimization. This can cause gradient explosion and degrade model performance. To address this, LINE samples edges with probabilities proportional to their weights and treats the sampled edges as binary during model updates.
Considerations
- The LINE algorithm treats all edges as undirected, ignoring their original direction.
Example Graph
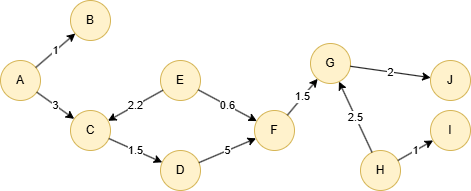
Run the following statements on an empty graph to define its structure and insert data:
ALTER EDGE default ADD PROPERTY {
score float
};
INSERT (A:default {_id: "A"}),
(B:default {_id: "B"}),
(C:default {_id: "C"}),
(D:default {_id: "D"}),
(E:default {_id: "E"}),
(F:default {_id: "F"}),
(G:default {_id: "G"}),
(H:default {_id: "H"}),
(I:default {_id: "I"}),
(J:default {_id: "J"}),
(A)-[:default {score: 1}]->(B),
(A)-[:default {score: 3}]->(C),
(C)-[:default {score: 1.5}]->(D),
(D)-[:default {score: 5}]->(F),
(E)-[:default {score: 2.2}]->(C),
(E)-[:default {score: 0.6}]->(F),
(F)-[:default {score: 1.5}]->(G),
(G)-[:default {score: 2}]->(J),
(H)-[:default {score: 2.5}]->(G),
(H)-[:default {score: 1}]->(I);
create().edge_property(@default, "score", float);
insert().into(@default).nodes([{_id:"A"},{_id:"B"},{_id:"C"},{_id:"D"},{_id:"E"},{_id:"F"},{_id:"G"},{_id:"H"},{_id:"I"},{_id:"J"}]);
insert().into(@default).edges([{_from:"A", _to:"B", score:1}, {_from:"A", _to:"C", score:3}, {_from:"C", _to:"D", score:1.5}, {_from:"D", _to:"F", score:5}, {_from:"E", _to:"C", score:2.2}, {_from:"E", _to:"F", score:0.6}, {_from:"F", _to:"G", score:1.5}, {_from:"G", _to:"J", score:2}, {_from:"H", _to:"G", score:2.5}, {_from:"H", _to:"I", score:1}]);
Creating HDC Graph
To load the entire graph to the HDC server hdc-server-1 as my_hdc_graph:
CREATE HDC GRAPH my_hdc_graph ON "hdc-server-1" OPTIONS {
nodes: {"*": ["*"]},
edges: {"*": ["*"]},
direction: "undirected",
load_id: true,
update: "static"
}
hdc.graph.create("my_hdc_graph", {
nodes: {"*": ["*"]},
edges: {"*": ["*"]},
direction: "undirected",
load_id: true,
update: "static"
}).to("hdc-server-1")
Parameters
Algorithm name: line
Name |
Type |
Spec |
Default |
Optional |
Description |
|---|---|---|---|---|---|
edge_schema_property |
[]"@<schema>?.<property>" |
/ | / | No | Specifies numeric edge properties used as weights by summing their values. Only properties of numeric type are considered, and edges without these properties are ignored. |
dimension |
Integer | ≥2 | / | No | Dimensionality of the embeddings. |
train_total_num |
Integer | ≥1 | / | No | Total number of training iterations. |
train_order |
Integer | 1, 2 |
1 |
Yes | Type of proximity to preserve, 1 means first-order proximity, 2 means second-order proximity. |
learning_rate |
Float | (0,1) | / | No | Learning rate used initially for training the model, which decreases after each training iteration until reaches min_learning_rate. |
min_learning_rate |
Float | (0,learning_rate) |
/ | No | Minimum threshold for the learning rate as it is gradually reduced during the training. |
neg_num |
Integer | ≥1 | / | Yes | Number of negative samples to produce for each positive sample, it is suggested to set between 1 to 10. |
resolution |
Integer | ≥1 | 1 |
Yes | The parameter used to enhance negative sampling efficiency; a higher value offers a better approximation to the original noise distribution; it is suggested to set as 10, 100, etc. |
return_id_uuid |
String | uuid, id, both |
uuid |
Yes | Includes _uuid, _id, or both values to represent nodes in the results. |
limit |
Integer | ≥-1 | -1 |
Yes | Limits the number of results returned. Set to -1 to include all results. |
File Writeback
CALL algo.line.write("my_hdc_graph", {
return_id_uuid: "id",
edge_schema_property: "score",
dimension: 3,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001,
neg_number: 5,
resolution: 100
}, {
file: {
filename: "embeddings"
}
})
algo(line).params({
projection: "my_hdc_graph",
return_id_uuid: "id",
edge_schema_property: "score",
dimension: 3,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001,
neg_number: 5,
resolution: 100
}).write({
file:{
filename: 'embeddings'
}
})
_id,embedding_result
J,0.134156,0.147224,-0.127542,
D,-0.110004,0.0445883,0.101143,
F,0.0306388,0.00733918,-0.11945,
H,0.0739559,-0.145214,0.0416526,
B,-0.02514,-0.0317756,-0.110371,
A,-0.0629673,0.0857267,0.100926,
E,0.0989685,0.0532481,0.0514933,
C,-0.127659,-0.136062,0.166552,
I,0.126709,0.103002,-0.0201314,
G,0.134589,-0.0157018,0.0237946,
DB Writeback
Writes the embedding_result values from the results to the specified node property. The property type is float[].
CALL algo.line.write("my_hdc_graph", {
return_id_uuid: "id",
edge_schema_property: "score",
dimension: 3,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001,
neg_number: 5,
resolution: 100
}, {
db: {
property: "vector"
}
})
algo(line).params({
projection: "my_hdc_graph",
return_id_uuid: "id",
edge_schema_property: "score",
dimension: 3,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001,
neg_number: 5,
resolution: 100
}).write({
db: {
property: "vector"
}
})
Full Return
CALL algo.line.run("my_hdc_graph", {
return_id_uuid: "id",
edge_schema_property: '@default.score',
dimension: 4,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001
}) YIELD embeddings
RETURN embeddings
exec{
algo(line).params({
return_id_uuid: "id",
edge_schema_property: '@default.score',
dimension: 4,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001
}) as embeddings
return embeddings
} on my_hdc_graph
Result:
_id |
embedding_result |
|---|---|
| J | [0.10039449483156204,0.11040361225605011,-0.095774807035923,-0.0819125771522522] |
| D | [0.0338507741689682,0.07575879991054535,0.023179633542895317,0.005116916261613369] |
| F | [-0.08940199762582779,0.05569583922624588,-0.10888427495956421,0.03145558759570122] |
| H | [-0.018580902367830276,-0.024093549698591232,-0.08332693576812744,-0.04651310294866562] |
| B | [0.0645807683467865,0.07539491355419159,0.07396885752677917,0.040248531848192215] |
| A | [0.03878217190504074,-0.09487584978342056,-0.10220742225646973,0.12434131652116776] |
| E | [0.09489674866199493,0.07733562588691711,-0.015262083150446415,0.10043458640575409] |
| C | [-0.01225052960216999,0.018636001273989677,-0.0036492166109383106,-0.0745576024055481] |
| I | [0.0327218696475029,-0.033543504774570465,-0.09600748121738434,0.11864277720451355] |
| G | [0.1094101220369339,-0.02683556079864502,-0.006420814897865057,0.012586096301674843] |
Stream Return
CALL algo.line.stream("my_hdc_graph", {
return_id_uuid: "id",
edge_schema_property: '@default.score',
dimension: 3,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001
}) YIELD embeddings
RETURN embeddings
exec{
algo(line).params({
return_id_uuid: "id",
edge_schema_property: '@default.score',
dimension: 3,
train_total_num: 10,
train_order: 1,
learning_rate: 0.01,
min_learning_rate: 0.0001
}).stream() as embeddings
return embeddings
} on my_hdc_graph
Result:
_id |
embedding_result |
|---|---|
| J | [0.13375547528266907,0.14727242290973663,-0.12761451303958893] |
| D | [-0.10898537188768387,0.045617908239364624,0.09961149096488953] |
| F | [0.030374202877283096,0.0074587357230484486,-0.11920187622308731] |
| H | [0.0746561735868454,-0.1447625607252121,0.04158430173993111] |
| B | [-0.02504475973546505,-0.0319061279296875,-0.1105244979262352] |
| A | [-0.0629904493689537,0.08570709079504013,0.1011749655008316] |
| E | [0.09852229058742523,0.0527675524353981,0.05207677558064461] |
| C | [-0.1265311986207962,-0.1361672431230545,0.16603653132915497] |
| I | [0.12704375386238098,0.10234453529119492,-0.0199428740888834] |
| G | [0.1342623233795166,-0.016277270391583443,0.0242084339261055] |

